For you ancestral-origin addicts, if your kit isn’t already uploaded to Gedmatch, you might want to consider adding it to their free database. Not only can you get additional matches from the Big Three DNA companies, but there are numerous admixture tools to explore, including Vadim Verenich’s new MDLP K13 Ultimate.
Vadim discusses and explains his methodology in creating MDLP K13 Ultimate here. According to Vadim, the tool is especially useful for showing the deep ancestry of Western Europeans.
MDLP K13 Ultimate’s Components
- Amerindian – the modal component of the Native American
- ANE – the modal component of the Northern Eurasians, which has been isolated from the common cluster with WHG – the highest values in the samples of MA1, AG2, as well as the ancient genomes from Sintashta, Andronov, Afanasievo, Yamnaya, Corded Ware etc. Among the modern populations the highest percentage of ANE has been detected in Kalash population. Almost the same with the ANE component in Lazaridis et al. 2014
- Arctic – modal component with peak populations Koryak, Chukchi, Eskimos and Itelmens
- ASI – еру modal component of South Indian populations (i assume that this component is identical to ASI in (Reich et al. 2009).
- Caucasus-Gedrosia – identical to Pontikos’s Caucasus-Gedrosia cluster
- EastAsian – the modal component of East Asia
- ENF – the component of the ancient European Neolithic farmers with the peak in the ancient samples of LBK culture (Lazaridis et al. 2014, Haak et al. 2015). Among the modern populations – the highest values have been detected in Sardinians, Corsicans and Basques.
- NearEast – the modal component of Middle Easterners
- Oceanian – the modal component of the aboriginal inhabitants of Oceania, Austronesian, Melanesia and Micronesia (the peak in modern Papuans and Australian Aborigines)
- Paleo-African – the modal component of African Pygmies and Bushmen
- Siberian – the modal component of south eastern Siberia
- Subsaharian – the second African component (Mandinka, Yoruba and Esan)
- WHG-UHG – the native component of the ancient European Mesolithic hunter-gatherers (Lazaridis et al. 2014, Haak et al. 2015). Among the modern populations – the highest percentage in the population of Estonians, Lithuanians, Finns and others.
A Test Run
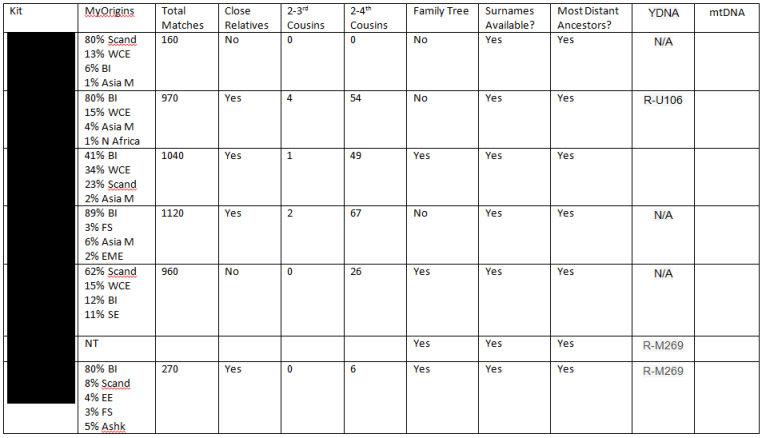
The testers in this experiment include 3 family groups that are predominantly Western European.
Group 1
Group 1 consists of 4 generations of my immediate family as shown in Figure 1.
Group 1 MDLP K13 Ultimate Admixture Results:
Oracles
Below are each tester’s Oracle 4 results. Oracles are designed to find the population(s) you are most similar to. As an example of what you might see in your own results, for Gladys I have shown the full Oracle 4 output. For the rest of the testers, I’ve only included the top estimate in each of the 4 population approximations. When using the Oracles, ideally you want your results to be a distance close to 1 or less. The further the distance, the less your sample matches the reference population. Please note, the MDLP K13 Ultimate Calculator has no Irish samples and thus no Irish population is included in the Oracle estimates.
Gladys mtDNA J1c3e Irish, small amount Alsace
Using 1 population approximation:
1 Germany_South @ 4.725954
2 Welsh @ 4.760169
3 Hungary @ 5.075449
4 Slovenian2 @ 5.144983
5 Slovak @ 5.262948
6 Austria @ 5.263268
7 North_European @ 5.285005
8 Czech2 @ 5.398787
9 Belgian @ 5.461259
10 German @ 5.465462
11 Slovenian @ 5.676105
12 South-German @ 5.796731
13 Austrian @ 5.843022
14 Hungarian @ 5.923850
15 Germany_North @ 6.175276
16 Inkeri @ 6.573199
17 Croat_BH @ 6.697131
18 North_German @ 6.715919
19 English_GBR @ 6.953663
20 Moldavian @ 7.103191
Using 2 populations approximation:
1 50% France +50% Vepsa @ 3.396806
Using 3 populations approximation:
1 50% Icelandic +25% Lak +25% Spanish_Pais_Vasco_IBS @ 2.251328
Using 4 populations approximation:
1 Avar + Basque_French + Orcadian + Swedish @ 1.562552
2 Avar + Basque_Spanish + Norwegian + Swedish @ 1.587619
3 Avar + Basque_French + Norwegian + Swedish @ 1.589907
4 Basque_Spanish + Lak + Norwegian + Swedish @ 1.604312
5 Avar + Basque_French + Icelandic + Swedish @ 1.609077
6 Avar + Basque_Spanish + Orcadian + Swedish @ 1.617271
7 Avar + Basque_Spanish + Scottish_Argyll_Bute_GBR + Swedish @ 1.627082
8 Basque_Spanish + Icelandic + Lak + Swedish @ 1.632314
9 Basque_French + Icelandic + Lak + Swedish @ 1.636813
10 Avar + French_South + Orcadian + Swedish @ 1.652004
11 Basque_Spanish + Norwegian + Swedish + Tabasaran @ 1.676389
12 Basque_French + Lak + Norwegian + Swedish @ 1.702851
13 Avar + Basque_Spanish + Icelandic + Swedish @ 1.707930
14 Basque_Spanish + Orcadian + Swedish + Tabasaran @ 1.720994
15 Avar + Basque_French + Scottish_Argyll_Bute_GBR + Swedish @ 1.737037
16 Avar + Basque_French + Orcadian + Russia @ 1.739913
17 Avar + French_South + Icelandic + Sweden @ 1.740096
18 Avar + Basque_Spanish + Russia + Scottish_Argyll_Bute_GBR @ 1.742547
19 Basque_Spanish + Icelandic + Swedish + Tabasaran @ 1.748318
20 Avar + Basque_Spanish + Orcadian + Russia @ 1.759932
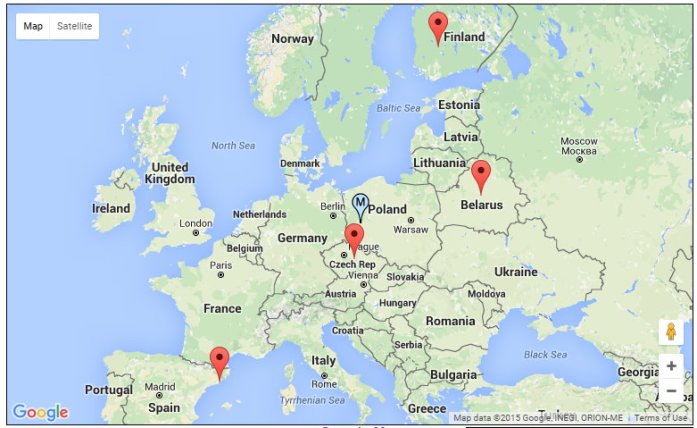
To give a visual representation, I have plotted her 4 population approximation with the Geographic Midpoint Calculator. The thumbtack with the ‘M’ is the midpoint between the 4 locations. At FTDNA, Gladys is 99% British Isles and 1% Asia Minor.
Steve mtDNA J1c3e yDNA R-Z251 ½ Irish, ½ East Flanders
Using 1 population approximation:
1 Germany_South @ 2.569169
Using 2 populations approximation:
1 50% Germany_South +50% Germany_South @ 2.569169
Using 3 populations approximation:
1 50% English_GBR +25% Greek_Comas +25% Vepsa @ 2.216009
Using 4 populations approximation:
1 Basque_Spanish + English_Kent_GBR + Kumyk_Stalskoe + Polish @ 1.199786
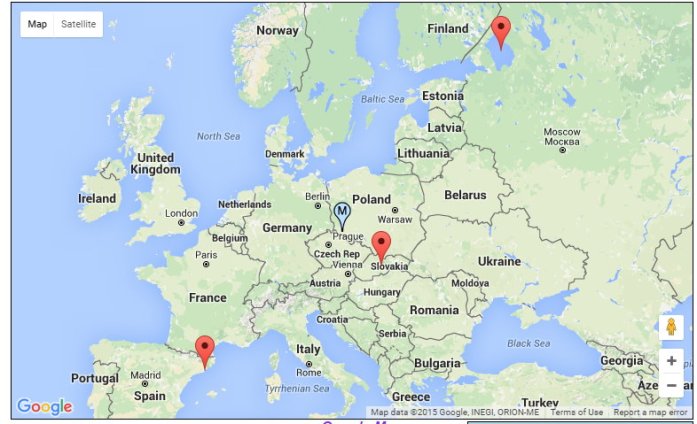
Steve’s midpoint plots in the Czech Republic. Although he is half Irish and half East Flemish, he also has small amounts of Asia Minor and Southern and Eastern European at FTDNA:
Lori mtDNA W3
Using 1 population approximation:
1 North_European @ 3.871830
Using 2 populations approximation:
1 50% French +50% Vepsa @ 3.446190
Using 3 populations approximation:
1 50% English_Kent_GBR +25% French_South +25% Tajik_Yagnobi @ 2.150152
Using 4 populations approximation:
1 Basque_Spanish + English_Kent_GBR + Orcadian + Tajik_Yagnobi @ 1.698324
Jeremy mtDNA W3 ¼ English (Newcastle), ¼ Colonial
Using 1 population approximation:
1 Welsh @ 1.196703
Using 2 populations approximation:
1 50% Welsh +50% Welsh @ 1.196703
Using 3 populations approximation:
1 50% English_Cornwall_GBR +25% English_GBR +25% Romanians @ 0.840597
Using 4 populations approximation:
1 English_Cornwall_GBR + English_Cornwall_GBR + English_GBR + Romanians @ 0.840597
Gavan mtDNA W3 half Irish and Scots
Using 1 population approximation:
1 English_GBR @ 3.039722
Using 2 populations approximation:
1 50% France +50% Scottish_Argyll_Bute_GBR @ 2.266037
Using 3 populations approximation:
1 50% English_GBR +25% Spanish_Aragon_IBS +25% Vepsa @ 1.150668
Using 4 populations approximation:
1 Belgian + English_Cornwall_GBR + Spanish_Aragon_IBS + Vepsa @ 1.059918
Leona mtDNA W3 ½ Pomeranian, ¼ Norwegian, ¼ Alsatian
Using 1 population approximation:
1 Welsh @ 1.911880
Using 2 populations approximation:
1 50% North European +50% Slovenian2 @ 1.408170
Using 3 populations approximation:
1 50% North_European +25% Slovak +25% Welsh @ 1.243836
Using 4 populations approximation:
1 French_South + Georgian + Latvian + Norwegian @ 1.187358
Jackie mtDNA W3
Using 1 population approximation:
1 Belgian @ 1.387411
Using 2 populations approximation:
1 50% Belgian +50% Welsh @ 1.096892
Using 3 populations approximation:
1 50% Belgian +25% Welsh +25% Welsh @ 1.096892
Using 4 populations approximation:
1 French_South + Georgian + Icelandic + Sorbs @ 1.075442
Diane
Using 1 population approximation:
1 South-German @ 1.934086
Using 2 populations approximation:
1 50% South-German +50% South-German @ 1.934086
Using 3 populations approximation:
1 50% North_German +25% Romanian +25% United-Kingdom @ 1.519436
Using 4 populations approximation:
1 English_Cornwall_GBR + Romanians + Slovak + United-Kingdom @ 1.441976
Group 2
MDLP K13 Ultimate Admixture Results
As seen in Figure 1, Group 2 includes 2 unrelated mothers, Nancy and Lori, and their sons, AJ and Jeremy who are ½ uncle and nephew.
Oracles
Nancy
Using 1 population approximation:
1 English_GBR @ 1.889876
Using 2 populations approximation:
1 50% English_GBR +50% English_GBR @ 1.889876
Using 3 populations approximation:
1 50% Russian_Orjol +25% Spanish_Pais_Vasco_IBS +25% Welsh @ 1.239205
Using 4 populations approximation:
1 Basque_French + Czech2 + Inkeri + West-Belarusian @ 1.045225
AJ
Using 1 population approximation:
1 North_European @ 2.072961
Using 2 populations approximation:
1 50% European_Utah +50% Welsh @ 1.128120
Using 3 populations approximation:
1 50% Czech2 +25% Spanish_Cantabria_IBS +25% Ukranian @ 0.942059
Using 4 populations approximation:
1 Czech2 + Slovak + Sorbs + Spanish_Cataluna_IBS @ 0.914081
Lori mtDNA W3
Using 1 population approximation:
1 North_European @ 3.871830
Using 2 populations approximation:
1 50% French +50% Vepsa @ 3.446190
Using 3 populations approximation:
1 50% English_Kent_GBR +25% French_South +25% Tajik_Yagnobi @ 2.150152
Using 4 populations approximation:
1 Basque_Spanish + English_Kent_GBR + Orcadian + Tajik_Yagnobi @ 1.698324
Jeremy mtDNA W3 ¼ English (Newcastle), ¼ Colonial
Using 1 population approximation:
1 Welsh @ 1.196703
Using 2 populations approximation:
1 50% Welsh +50% Welsh @ 1.196703
Using 3 populations approximation:
1 50% English_Cornwall_GBR +25% English_GBR +25% Romanians @ 0.840597
Using 4 populations approximation:
1 English_Cornwall_GBR + English_Cornwall_GBR + English_GBR + Romanians @ 0.840597
Group 3
MDLP K13 Ultimate Admixture Results
Group 3 consists of two full sisters, Jean and Sally, and Sally’s daughter Julie. Jean and Sally’s father was born in Glasgow from a family that originated primarily in the north of Scotland. Maternally, they derive mainly from early Irish and German immigrants. Paternally, Julie’s family has lived in Lippe, Germany for centuries.
Oracles
Jean 1/2 Northern Scots, ½ Colonial
Using 1 population approximation:
1 North_German @ 4.487704
Using 2 populations approximation:
1 50% English_Cornwall_GBR +50% Vepsa @ 3.162740
Using 3 populations approximation:
1 50% English_Cornwall_GBR +25% Scottish_Argyll_Bute_GBR +25% Vepsa @ 2.750026
Using 4 populations approximation:
1 English_Cornwall_GBR + English_Cornwall_GBR + Scottish_Argyll_Bute_GBR + Vepsa @ 2.750026
Sally ½ Northern Scots, half Colonial
Using 1 population approximation:
1 North_European @ 1.952304
Using 2 populations approximation:
1 50% English_GBR +50% North_European @ 1.668977
Using 3 populations approximation:
1 50% Slovak +25% Spanish_Pais_Vasco_IBS +25% Vepsa @ 1.005733
Using 4 populations approximation:
1 Basque_French + Hungary + Russian_Smolensk + Vepsa @ 0.987646
Sally has a small amount of Central Asia in her FTDNA ancestral origins that her sister Jean does not have.
Julie ¼ Northern Scots, ¼ Colonial, half German (Lippe)
Using 1 population approximation:
1 English_GBR @ 3.404993
Using 2 populations approximation:
1 50% French +50% Vepsa @ 3.007144
Using 3 populations approximation:
1 50% North_German +25% Spanish_Valencia_IBS +25% Vepsa @ 2.082997
Using 4 populations approximation:
1 Basque_French + Slovak + Vepsa + Vepsa @ 1.434730
Conclusion:
For my testers, the admixture calculators (ANR, ENF, WHG etc) appear reasonable and tick all the boxes I look for when considering the value of admixture tools. The Oracle estimates are another consideration. As the calculator is exposing deep ancestry, and as a whole very little of the group’s paper trail reaches the 1500’s, let alone precedes it, we have no way of determining its precision. We can, however, look at the population approximations passed down and across the group’s generations, as well as consistency between estimated regions and estimated regions/populations from other calculators.
Admittedly, I needed to google some of the reference populations to find where to plot them on the map. Despite the seemingly exotic populations some of our testers received in their estimates, once each 4 population approximation was plotted (and the non-tested parent’s known ancestry taken into account) for the most part each closely related family member plotted within the same general region.
Of course, as always, we must remember admixture calculators are estimates of ancestry, and a work in progress being improved upon as the science and reference samples grow. They are interesting, and as long as you don’t let unexpected admixture results derail your genetic and genealogical efforts, they can be entertaining.

















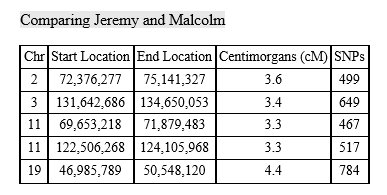











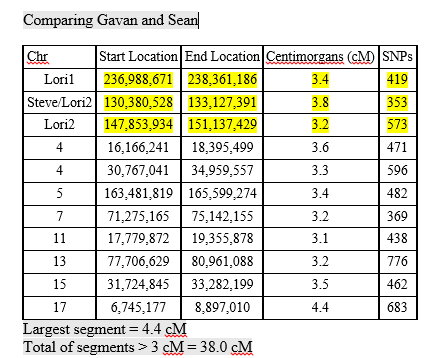
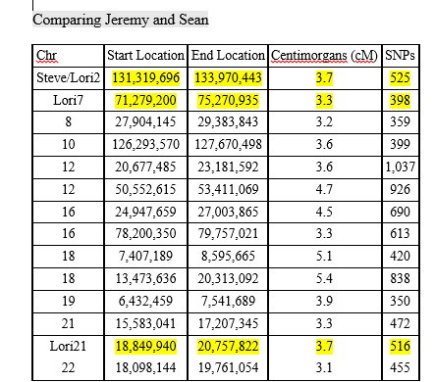





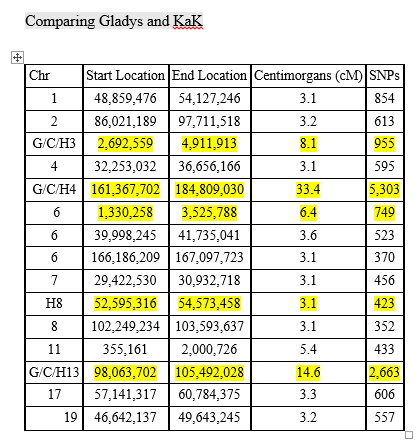
















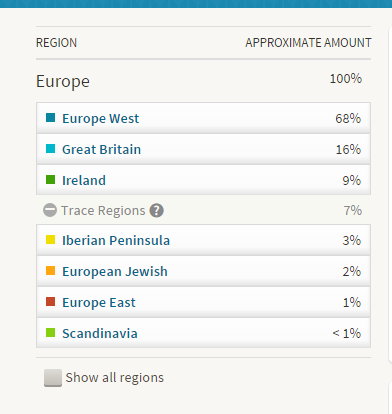








 With mostly recent and current European ancestry, my newly minted Ancestry DNA results only produced 11 Shared Ancestor hints. One of which, belonging to Rebecca, suggested our common ancestors are Michael Warren and Catherine Henton, my 7th great-grandparents. Ancestry classified us as 5th to 8th Cousins with confidence rated as, ‘Good.’
With mostly recent and current European ancestry, my newly minted Ancestry DNA results only produced 11 Shared Ancestor hints. One of which, belonging to Rebecca, suggested our common ancestors are Michael Warren and Catherine Henton, my 7th great-grandparents. Ancestry classified us as 5th to 8th Cousins with confidence rated as, ‘Good.’


 NA Project
NA Project